|
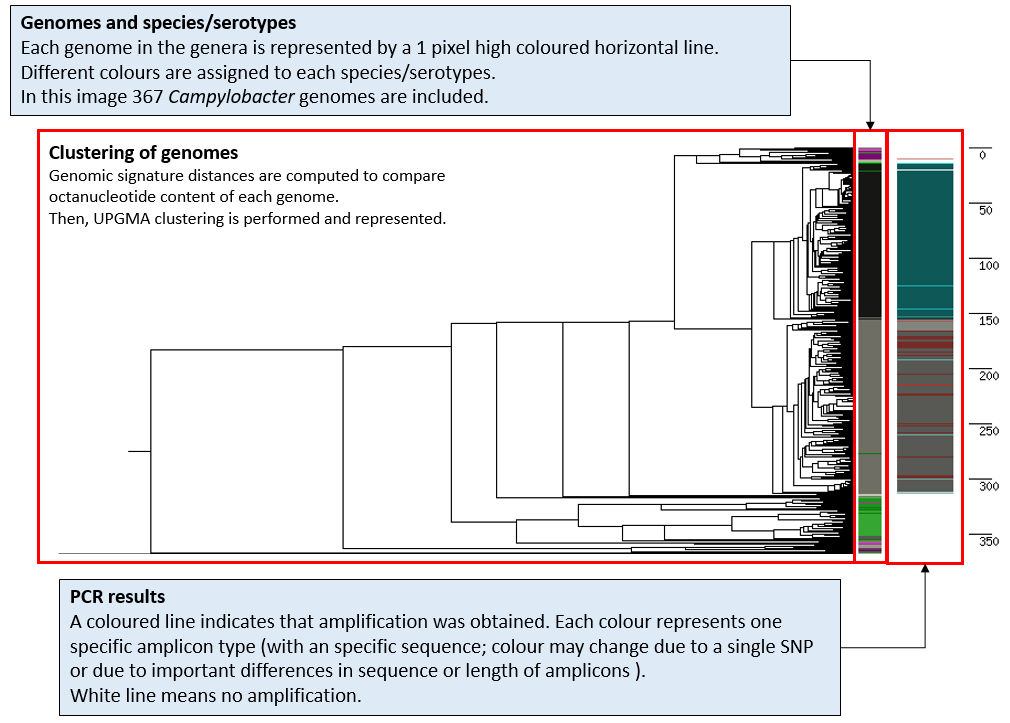
When computing PCR against all genomes within a bacterial genera,
amplification may be limited to specific species, serotypes or clusters.
Additionaly, the sequences or the PCR products are not always identical:
amplicons may differ in a single nucleotide, in several ones, or their lenght may differ.
In those cases, it is more likely to get identical PCR products in closely related genomes.
In order to easily visualice and detect the associations described above,
we are providing amplification results together with clusting information
for bacterial generas with over 200 genomes in our database.
To learn more about the clustering procedure, check here.
As an example, we simulated a PCR experiment with primers TTGGTATGGCTATAGGAACTCTTATAGCT and CACACCTGAAGTATGAAGTGGTCTAAGPGMA
described here to target
the Campylobacter jejuni-specific region of the ORF-C sequence. Amplicon types obtained in the experiments were included
within a dendrogram for all genomes in the genera. The diferent elements in the image are described:
All Campylobacter genomes included in the experiment are listed bellow.
The genomes are identified as ordered in the dendrogram and the amplicon(s) types are shown for each genome:
| 